All papers that have not been peer-reviewed will not appear here, including preprints. You can access my all of papers at 🔗Google Scholar.
2025

A Unified Framework for Multiple Instance Learning in Computational Pathology
Xitong Ling*, Minxi Ouyang*, Mengjia Li*, Tian Guan, Xiaoping Liu†, Yonghong He†(† corresponding author)
Github 2025 Repository
Computational pathology is emerging as a transformative diagnostic approach, leveraging AI to enable data-driven analysis of histopathological images. However, the ultra-high resolution of whole slide images (WSIs) challenges traditional image analysis methods. Multiple Instance Learning (MIL) addresses this by treating WSIs as weakly supervised "bags of instances," yet current MIL methods lack standardization, hindering reproducibility. To resolve this, we present MIL-BASELINE, a modular and extensible library for unified, reproducible MIL in computational pathology, supporting rapid prototyping and benchmarking.

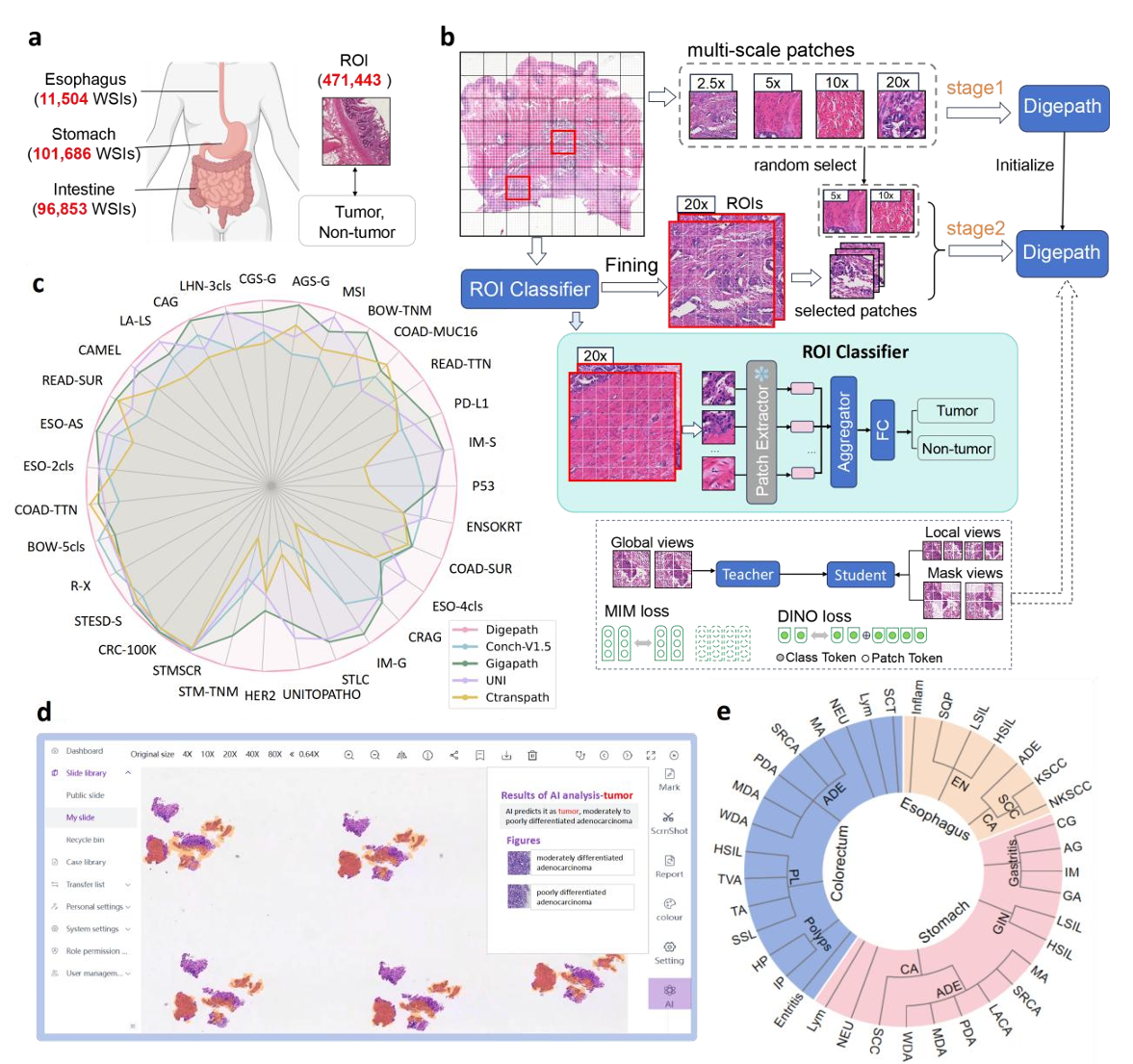
Subspecialty-Specific Foundation Model for Intelligent Gastrointestinal Pathology
Lianghui Zhu*, Xitong Ling*, Minxi Ouyang*, Xiaoping Liu, Mingxi Fu, Tian Guan, Fanglei Fu, Xuanyu Wang, Maomao Zeng, Mingxi Zhu, Yibo Jin, Liming Liu, Song Duan, Qiming He, Yizhi Wang, Luxi Xie†, Houqiang Li†, Sufang Tian†, Yonghong He†(† corresponding author)
Arxiv 2025 Journal
GI diseases pose a significant clinical burden, necessitating precise diagnostics. Conventional histopathology, relying on subjective pathologist interpretation, suffers from limited reproducibility. To address this and the lack of specialized pathology models for GI diseases, we developed Digepath, a GI pathology foundation model. It employs a dual-phase optimization strategy combining pretraining with fine-screening to detect sparse lesion areas in WSIs. Pretrained on over 353 million patches from 200,000 slides, Digepath achieves state-of-the-art performance on 33 out of 34 GI pathology tasks, including diagnosis, molecular prediction, and prognosis evaluation. It also demonstrates near-perfect sensitivity for early GI cancer screening across multiple institutions, highlighting its potential to enhance histopathological practice and serve as a paradigm for other pathology subspecialties.

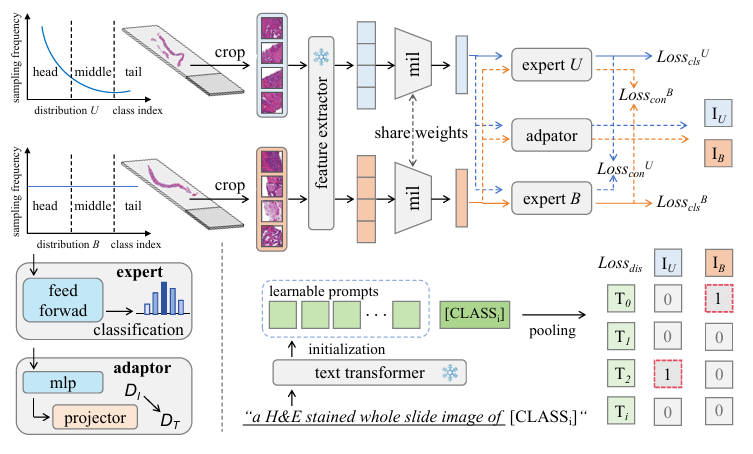
Multimodal Distillation-Driven Ensemble Learning for Long-Tailed Histopathology Whole Slide Images Analysis
Xitong Ling* Yifeng Ping* Jiawen Li* Jing Peng Yuxuan Chen Minxi Ouyang Yizhi Wang Yonghong He Tian Guan Xiaoping Liu†, († corresponding author)
Arxiv 2025 Conference
MIL is crucial in computational pathology for weakly supervised WSI analysis, but long-tailed distributions cause class imbalance issues. We propose an ensemble learning method with shared aggregators and consistency constraints to reduce class imbalance impact. Additionally, we introduce a multimodal distillation framework using pre-trained text encoders to enhance feature extraction. Our method, MDE-MIL, integrates multiple expert branches and achieves superior performance on Camelyon+-LT and PANDA-LT datasets.

A comprehensive foundation model for computational pathology with over 100 diverse clinical-grade tasks
Fang Yan*, Jianfeng Wu*, Jiawen Li*, Wei Wang, Jiaxuan Lu, Wen Chen, Zizhao Gao, Jianan Li, Hong Yan, Jiabo Ma, Minda Chen, Yang Lu, Qing Chen, Yizhi Wang, Xitong Ling, Xuenian Wang, Zihan Wang, Qiang Huang, Shengyi Hua, Mianxin Liu, Lei Ma, Tian Shen, Xiaofan Zhang†, Yonghong He†, Hao Chen†, Shaoting Zhang†, Zhe Wang†(† corresponding author)
Arxiv 2025 Journal
PathOrchestra, a pathology foundation model, addresses challenges in computational pathology by using self-supervised learning on a large dataset of 300K slides from 20 tissue types. It was evaluated on 112 clinical tasks, demonstrating strong performance in tasks like pan-cancer classification and biomarker assessment. PathOrchestra also explores multimodal tasks, including generating structured reports for colorectal cancer and lymphoma, showcasing its potential for clinical application.

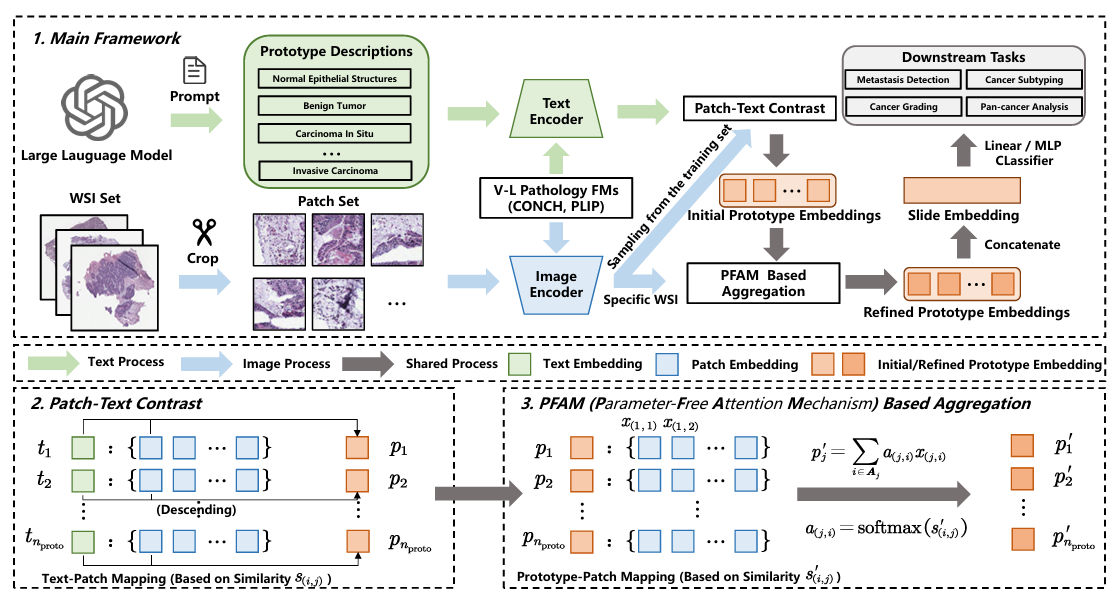
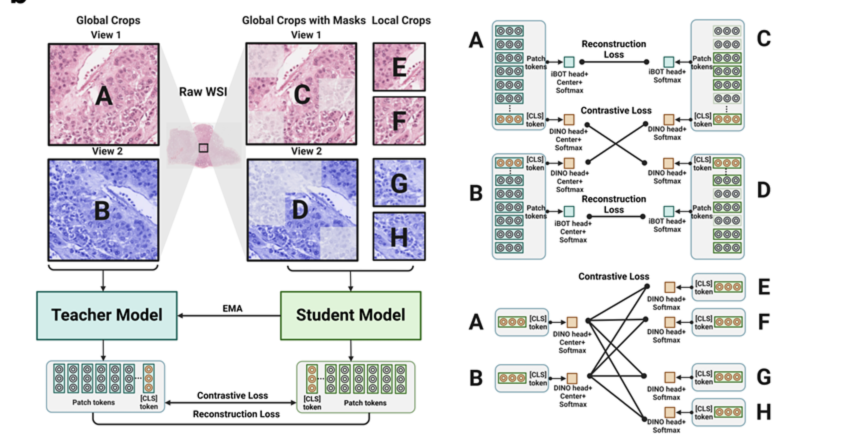
Unsupervised Slide Representation Learning via Patch-Text Contrast in Computational Pathology
Yuxuan Chen*, Jiawen Li*, Jiali Hu*, Xitong Ling, Tian Guan, Anjia Han†, Yonghong He†(† corresponding author)
Arxiv 2025 Conference
MIL is crucial in computational pathology for weakly supervised WSI analysis, but long-tailed distributions cause class imbalance issues. We propose an ensemble learning method with shared aggregators and consistency constraints to reduce class imbalance impact. Additionally, we introduce a multimodal distillation framework using pre-trained text encoders to enhance feature extraction. Our method, MDE-MIL, integrates multiple expert branches and achieves superior performance on Camelyon+-LT and PANDA-LT datasets.

2024
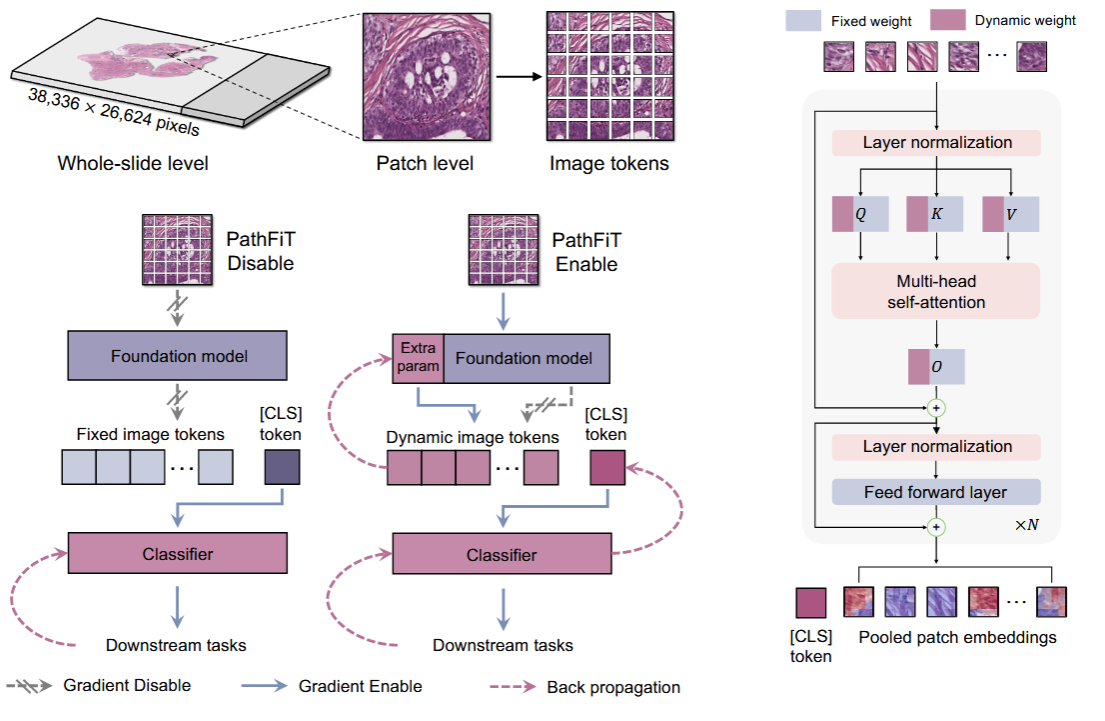
Unlocking adaptive digital pathology through dynamic feature learning
Jiawen Li*, Tian Guan*, Qingxin Xia, Yizhi Wang, Xitong Ling, Jing Li, Qiang Huang, Zihan Wang, Zhiyuan Shen, Yifei Ma, Zimo Zhao, Zhe Lei, Tiandong Chen, Junbo Tan, Xueqian Wang, Xiu-Wu Bian†, Zhe Wang†, Lingchuan Guo†, Chao He†, Yonghong He†(† corresponding author)
Arxiv 2024 Journal
Foundation models have transformed digital pathology by enabling quantitative analysis of histological patterns and cancer-specific signals. Yet, their static features limit adaptability to diverse clinical needs. We propose PathFiT, a plug-and-play dynamic feature learner that enhances the flexibility of pathology foundation models across varied tasks. To evaluate PathFiT, we build a 20TB benchmark covering 28 H&E-stained and 7 specialized imaging tasks. PathFiT achieves state-of-the-art results on 34 of 35 tasks, with notable gains on 23 and a 10.20% improvement on specialized imaging. These results highlight its strong adaptability and performance across digital pathology.

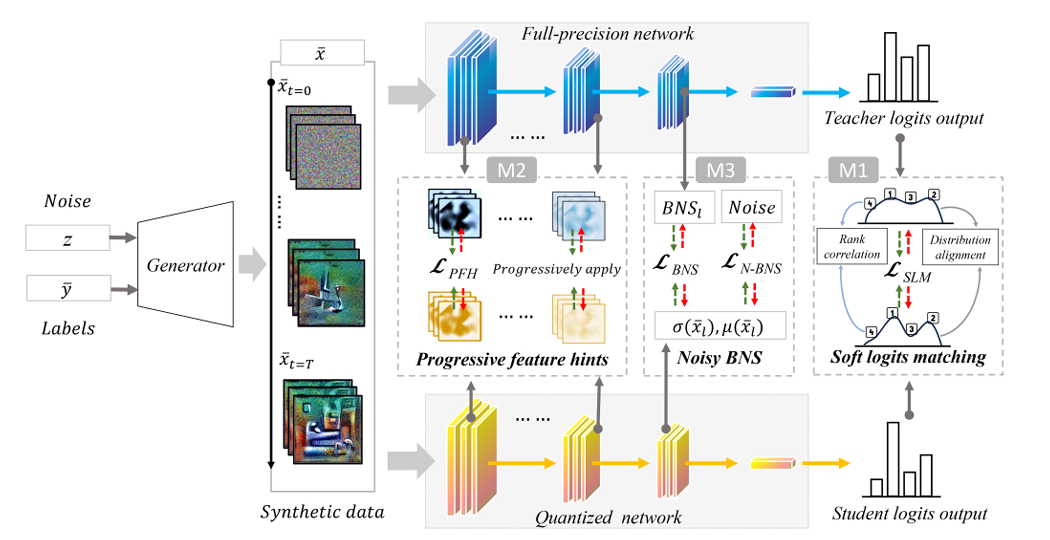
Low Bit-Width Zero-Shot Quantization With Soft Feature-Infused Hints for IoT Systems
Xinrui Chen*, Yizhi Wang*, Yao Li, Xitong Ling, Mengkui Li, Ruikang Liu, Minxi Ouyang, Kang Zhao, Tian Guan†, Yonghong He†(† corresponding author)
IEEE Internet of Things Journal 2024 Journal
Quantization enables deep learning on IoT devices by compressing neural networks, but most methods require private training data. Zero-shot quantization (ZSQ) addresses this by quantizing without access to training data. However, low-bit-width ZSQ faces challenges like hard logits matching, unstable feature alignment, and low synthetic data diversity. This article introduces S-ZSQ, a novel ZSQ framework that enhances knowledge transfer and synthetic data generation, enabling low-bit-width quantized networks to learn more effectively from full-precision networks. It achieves significant improvements on CIFAR-10/100 and ImageNet-1k with fewer fine-tuning epochs. For example, it outperforms AdaDFQ by 8.08%/11.16% in top-1 accuracy on 3-bit ResNet-18/ResNet-50 cases.

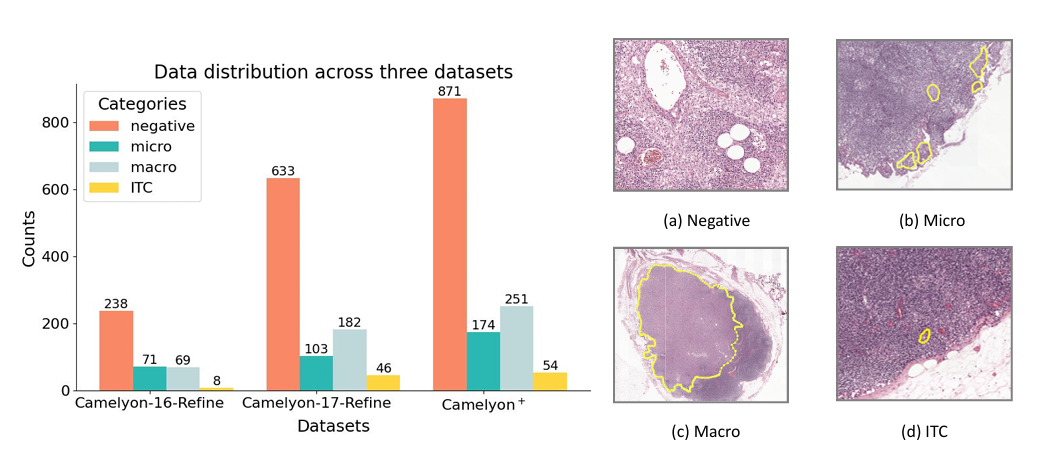
Towards a comprehensive benchmark for pathological lymph node metastasis in breast cancer sections
Xitong Ling*, Yuanyuan Lei*, Jiawen Li*, Junru Cheng, Wenting Huang, Tian Guan, Jian Guan†, Yonghong He†(† corresponding author)
Scientific Data 2024 Journal
Optical microscopy advancements have enabled whole slide imaging (WSI), facilitating AI-powered computational pathology (CPath). The Camelyon datasets are widely used benchmarks, yet label quality and clinical relevance remain underexplored. In this study, we reprocessed 1,399 WSIs from Camelyon-16 and -17, removing low-quality slides, correcting labels, and adding expert annotations to the test set. We refined the binary cancer task into a four-class classification: negative, ITC, micro-, and macro-metastasis. Using this cleaned dataset, we re-evaluated foundational models and MIL methods, offering a stronger benchmark for AI in histopathology.

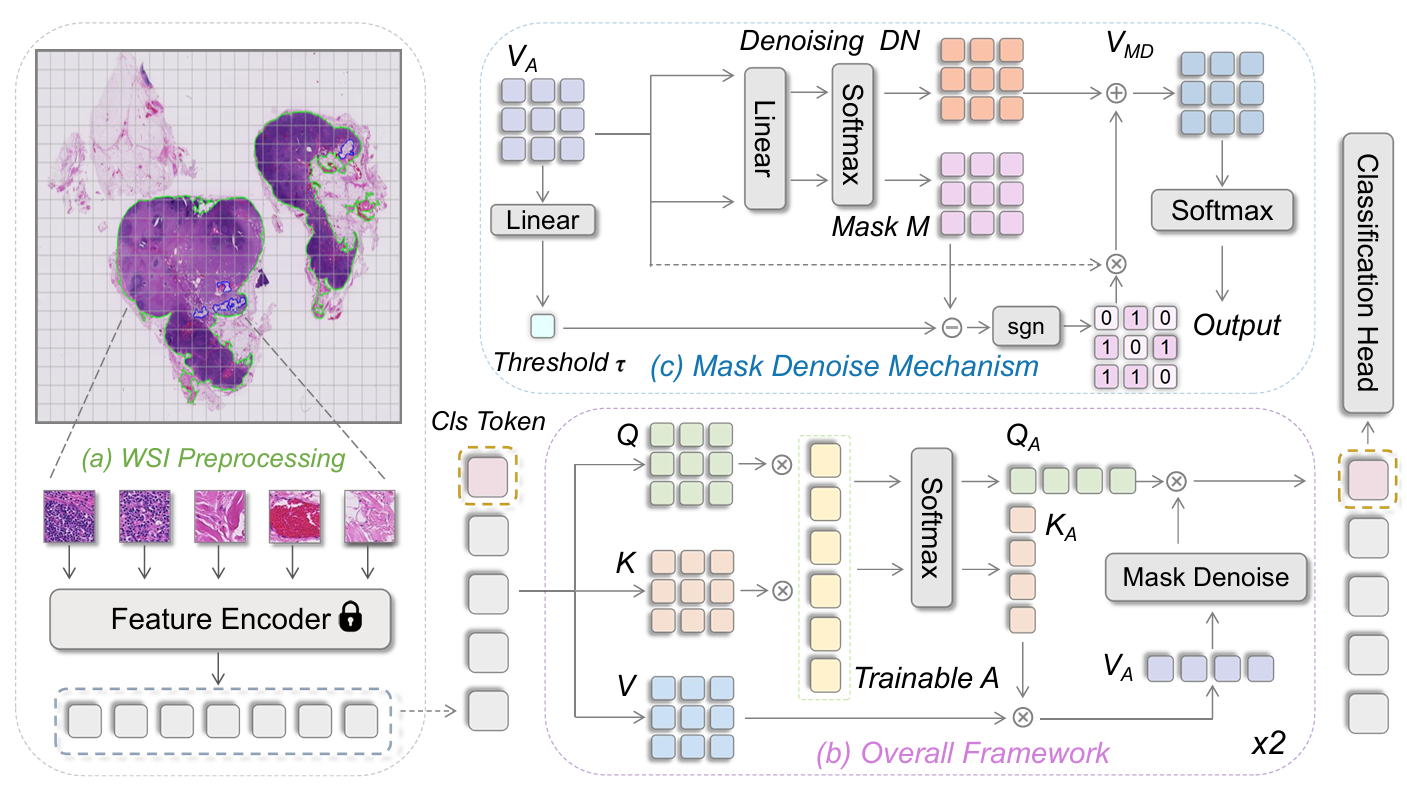
Agent Aggregator with Mask Denoise Mechanism for Histopathology Whole Slide Image Analysis
Xitong Ling*, Minxi Ouyang*, Yizhi Wang, Xinrui Chen, Renao Yan, Junru Cheng, Tian Guan, Sufang Tian, Xiaoping Liu†, Yonghong He†(† corresponding author)
ACM-MM 2024 ConferencePoster
Whole Slide Image (WSI) classification in histopathology faces challenges due to gigapixel resolution and lack of fine-grained annotations. Existing MIL-based attention mechanisms often neglect inter-instance relations and suffer from high complexity. We propose AMD-MIL, an agent-based aggregator with a mask denoise mechanism, which dynamically filters noise and enhances critical feature representations. Experiments on CAMELYON and TCGA datasets demonstrate superior performance and interpretability.

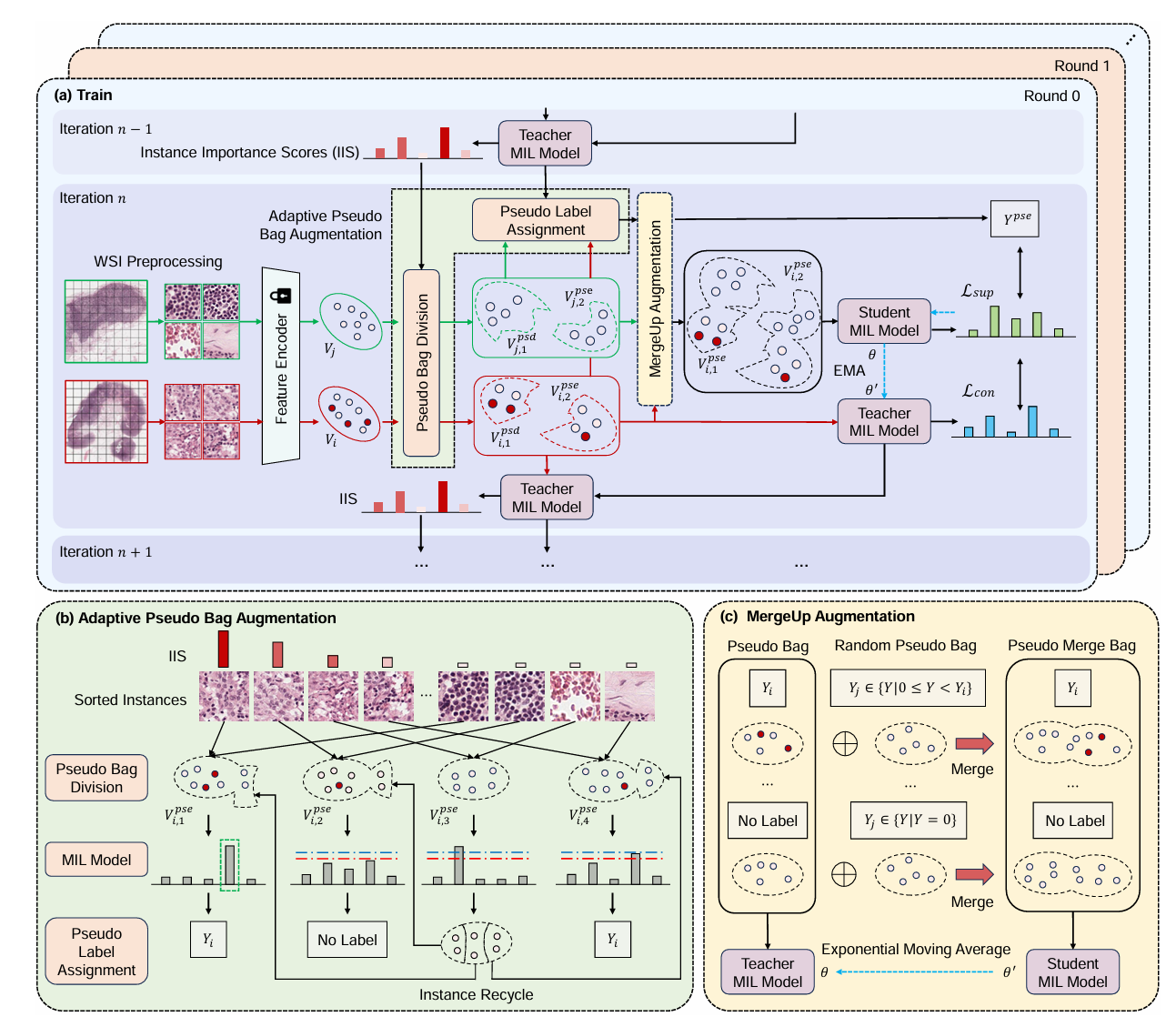
MergeUp-augmented Semi-Weakly Supervised Learning for WSI Classification
Mingxi Ouyang*, Yuqiu Fu*, Renao Yan*, ShanShan Shi, Xitong Ling, Lianghui Zhu†, Yonghong He†, Tian Guan†(† corresponding author)
Arxiv 2024 Conference
Recent progress in computational pathology and AI has enhanced WSI classification. However, the high resolution of WSIs and limited manual annotations pose challenges. MIL is a promising weakly supervised learning method for WSI classification. Research shows that pseudo bag augmentation can improve model performance by encouraging diverse data learning. Yet, directly using parent labels can introduce noise through mislabeling. To address this, we introduce SWS-MIL, which uses adaptive pseudo bag augmentation (AdaPse) to label data based on a threshold strategy. Additionally, we employ a "student-teacher" pattern with MergeUp, a feature augmentation technique that merges low-priority bags to enhance inter-category information and data diversity. Experiments on CAMELYON-16, BRACS, and TCGA-LUNG datasets demonstrate that our method outperforms current state-of-the-art approaches, confirming its effectiveness in WSI classification.
